Weights¶
In this notebook we demonstrate all of netCDF-SCM’s known weightings. These weights are used when taking area overages for different SCM boxes e.g. the ocean/land boxes or the El Nino box.
Note: here we use the “last resort” land surface fraction values. However, if land surface fraction data is available then that is used to do land/ocean weighting rather than the “last resort” values.
This notebook is set out as follows:
we show the default weights
we show how the different available options for combining area and surface fraction information
we show all our inbuilt weights
we show how the user can define their own custom weights.
Imports¶
[1]:
# NBVAL_IGNORE_OUTPUT
from os.path import join
import numpy as np
import iris
import iris.quickplot as qplt
import matplotlib.pyplot as plt
from netcdf_scm.iris_cube_wrappers import CMIP6OutputCube
from netcdf_scm.weights import (
AreaSurfaceFractionWeightCalculator,
AreaWeightCalculator,
get_weights_for_area,
WEIGHTS_FUNCTIONS_WITHOUT_AREA_WEIGHTING,
)
[2]:
plt.style.use("bmh")
%matplotlib inline
Data path¶
Here we use our test data.
[3]:
DATA_PATH_TEST = join("..", "..", "..", "tests", "test-data")
DATA_PATH_TEST_CMIP6_ROOT = join(DATA_PATH_TEST, "cmip6output")
Load the cube¶
[4]:
tas = CMIP6OutputCube()
tas.load_data_in_directory(
join(
DATA_PATH_TEST_CMIP6_ROOT,
"CMIP6/ScenarioMIP/BCC/BCC-CSM2-MR/ssp126/r1i1p1f1/Amon/tas/gn/v20190314",
)
)
Weights¶
Default weights¶
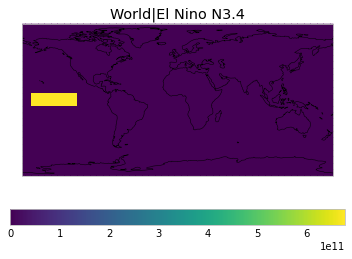
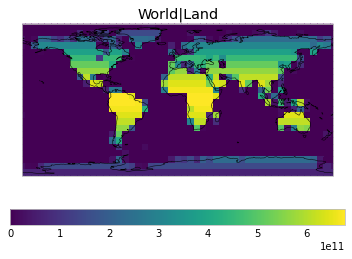
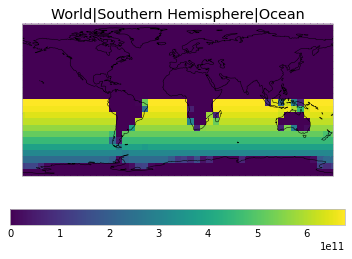
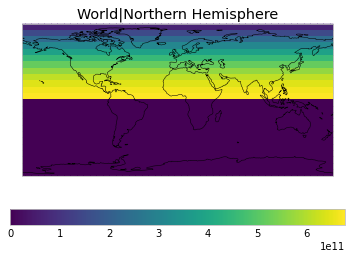
By default, only land/ocean and hemispheric weights are considered.
[5]:
# NBVAL_IGNORE_OUTPUT
default_weights = tas.get_scm_timeseries_weights()
/home/jared/miniconda3/envs/netcdf-scm-v2/lib/python3.8/site-packages/iris/analysis/cartography.py:394: UserWarning: Using DEFAULT_SPHERICAL_EARTH_RADIUS.
warnings.warn("Using DEFAULT_SPHERICAL_EARTH_RADIUS.")
/home/jared/miniconda3/envs/netcdf-scm-v2/lib/python3.8/site-packages/iris/analysis/cartography.py:394: UserWarning: Using DEFAULT_SPHERICAL_EARTH_RADIUS.
warnings.warn("Using DEFAULT_SPHERICAL_EARTH_RADIUS.")
[6]:
# NBVAL_IGNORE_OUTPUT
def plot_weights(weights_to_plot, constraint=None, axes=None, **kwargs):
for i, (label, weights) in enumerate(weights_to_plot.items()):
if axes is None:
ax = plt.figure().add_subplot(111)
else:
ax = axes[i]
weight_cube = tas.cube.collapsed("time", iris.analysis.MEAN)
weight_cube.data = weights
weight_cube.units = ""
if constraint is not None:
weight_cube = weight_cube.extract(constraint)
plt.sca(ax)
qplt.pcolormesh(
weight_cube, **kwargs,
)
plt.gca().set_title(label)
plt.gca().coastlines()
plot_weights(default_weights)
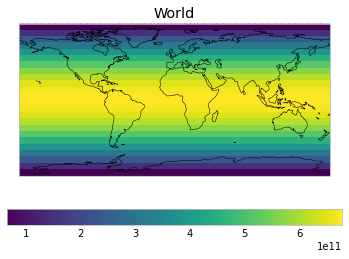
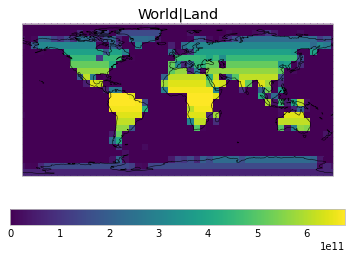
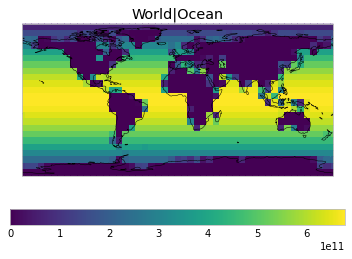
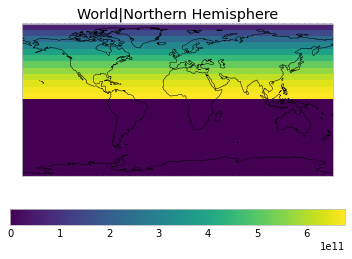
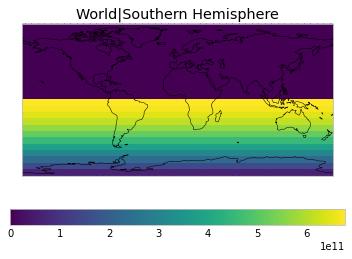
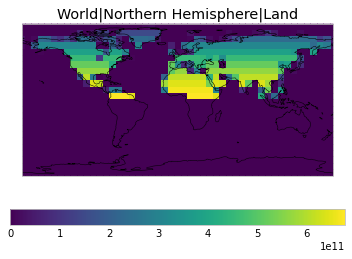
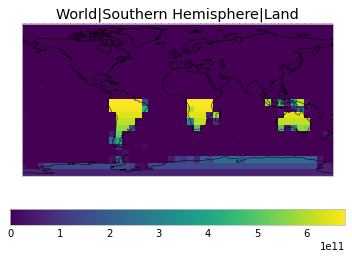
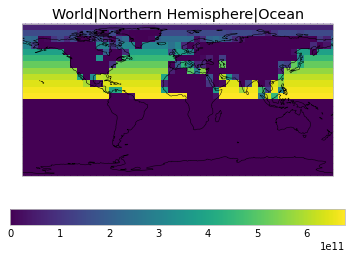
Area and surface fraction combination options¶
By defaults, the weights are calculated as the combination of area and surface fractions using netcdf_scm.weights.AreaSurfaceFractionWeightCalculator.
[7]:
# NBVAL_IGNORE_OUTPUT
print(AreaSurfaceFractionWeightCalculator.__doc__)
Calculates weights which are both area and surface fraction weighted
.. math::
w(lat, lon) = a(lat, lon) \\times s(lat, lon)
where :math:`w(lat, lon)` is the weight of the cell at given latitude and
longitude, :math:`a` is area of the cell and :math:`s` is the surface
fraction of the cell (e.g. fraction of ocean area for ocean based regions).
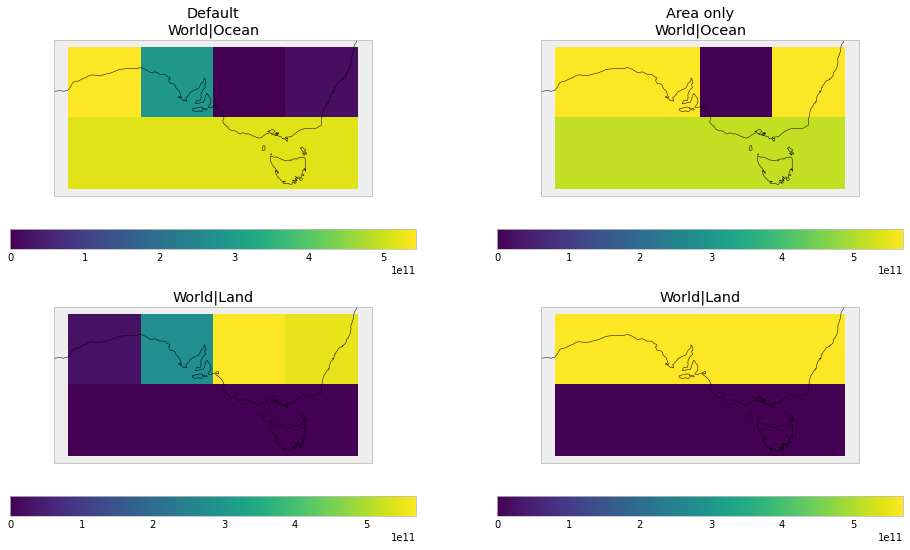
For land/ocean weights, this causes regions on coastlines to have weights less than their area weight, because they are not fully land or ocean.
The user can instead use netcdf_scm.weights.AreaWeightCalculator, which focusses on area weights but removes any areas that have a surface fraction of zero.
[8]:
# NBVAL_IGNORE_OUTPUT
print(AreaWeightCalculator.__doc__)
Calculates weights which are area weighted but surface fraction aware.
This means that any cells which have a surface fraction of zero will
receive zero weight, otherwise cells are purely area weighted.
.. math::
w(lat, lon) = \\begin{cases}
a(lat, lon), & s(lat, lon) > 0 \\\\
0, & s(lat, lon) = 0
\\end{cases}
where :math:`w(lat, lon)` is the weight of the cell at given latitude and
longitude, :math:`a` is area of the cell and :math:`s` is the surface
fraction of the cell (e.g. fraction of ocean area for ocean based regions).
[9]:
# NBVAL_IGNORE_OUTPUT
area_weights = tas.get_scm_timeseries_weights(cell_weights="area-only")
/home/jared/miniconda3/envs/netcdf-scm-v2/lib/python3.8/site-packages/iris/analysis/cartography.py:394: UserWarning: Using DEFAULT_SPHERICAL_EARTH_RADIUS.
warnings.warn("Using DEFAULT_SPHERICAL_EARTH_RADIUS.")
[10]:
# NBVAL_IGNORE_OUTPUT
fig, axes = plt.subplots(figsize=(16, 9), nrows=2, ncols=2)
for i, (w, title) in enumerate(
((default_weights, "Default"), (area_weights, "No land fraction"))
):
plt_weights = {k: w[k] for k in ["World|Ocean", "World|Land"]}
zoom_constraint = iris.Constraint(
latitude=lambda cell: -45 < cell < -25
) & iris.Constraint(longitude=lambda cell: 120 < cell < 160)
plot_weights(
plt_weights, constraint=zoom_constraint, axes=[axes[0][i], axes[1][i]],
)
cf = plt.gcf()
for i, (w, title) in enumerate(
((default_weights, "Default"), (area_weights, "Area only"))
):
title_ax = cf.axes[i * 4]
title_ax.set_title("{}\n{}".format(title, title_ax.get_title()))
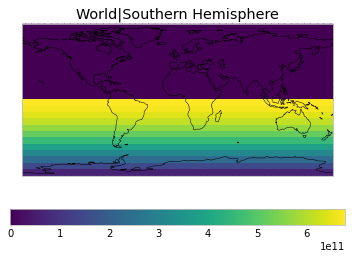
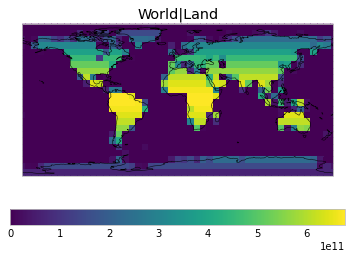
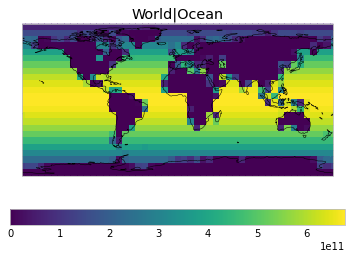
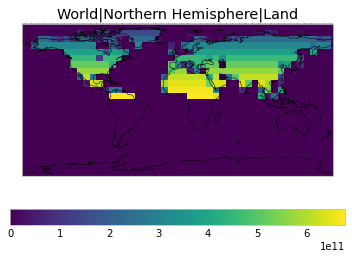
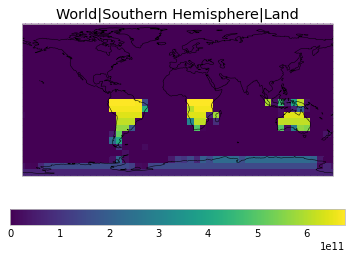
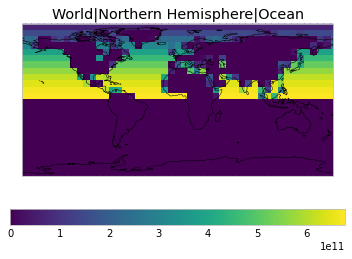
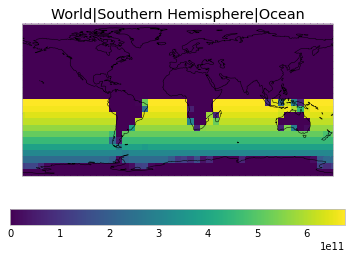
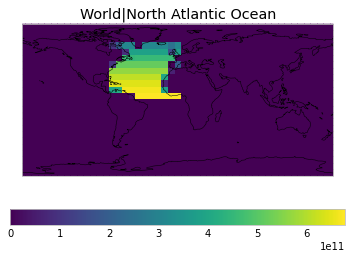
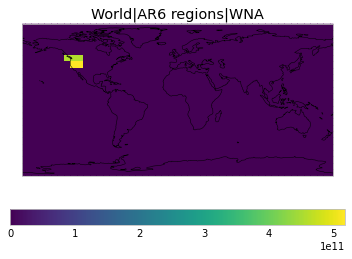
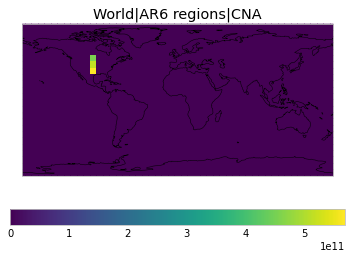
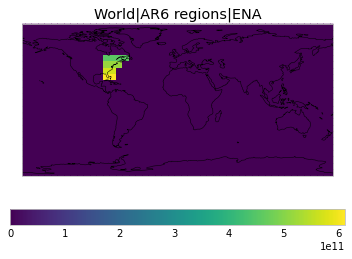
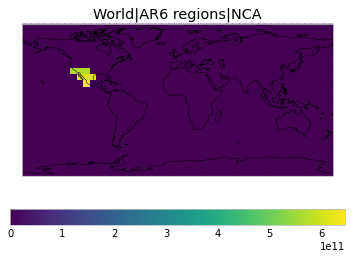
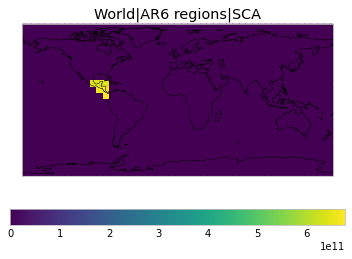
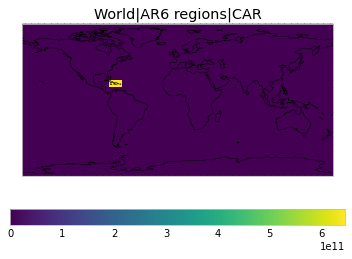
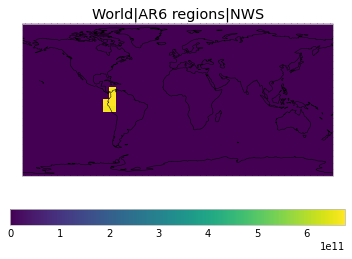
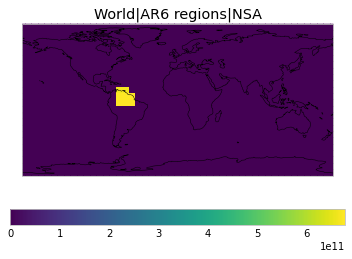
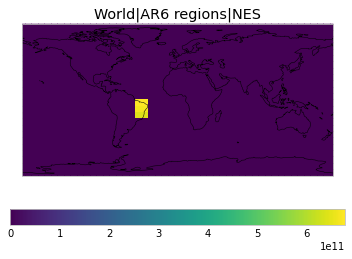
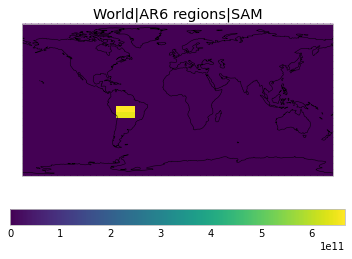
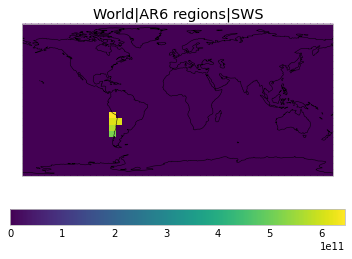
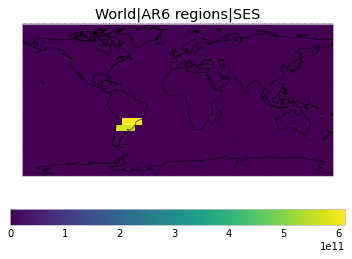
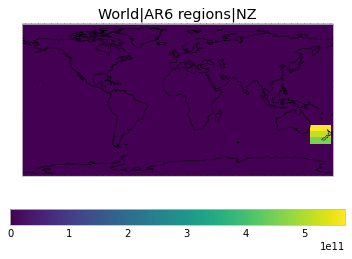
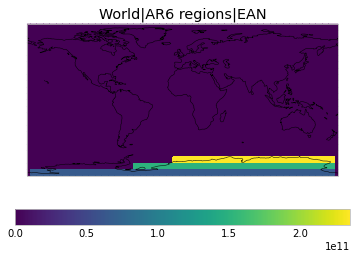
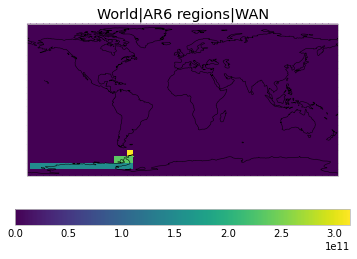
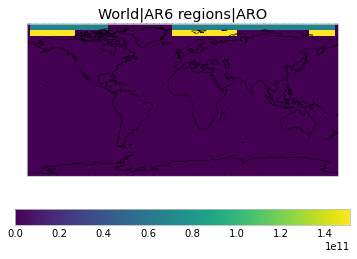
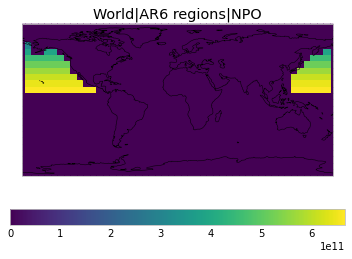
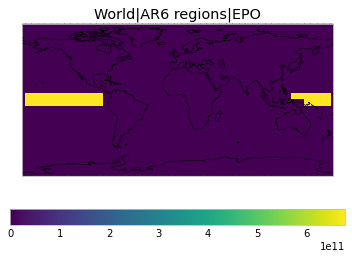
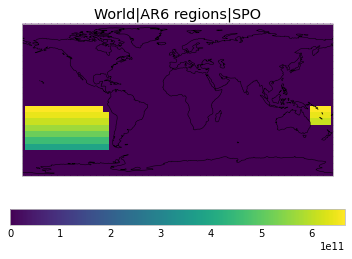
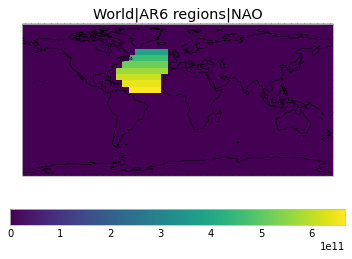
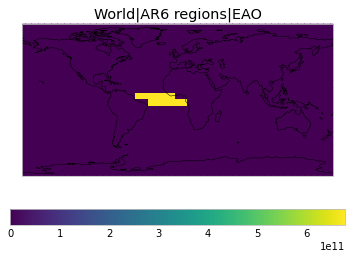
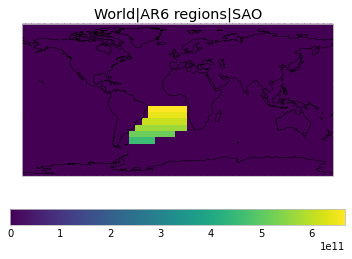
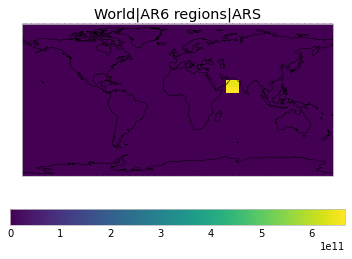
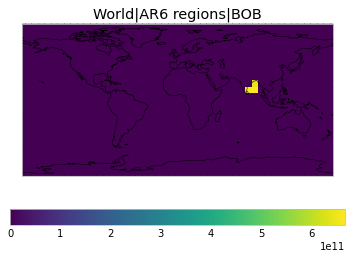
All inbuilt masks¶
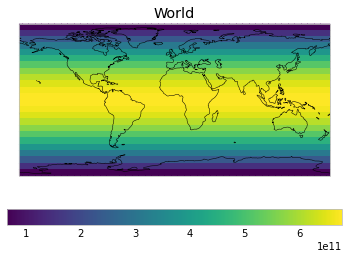
The default masks do not contain all inbuilt masks. We can use all available inbuilt masks instead.
These masks include 58 regions which will be used in IPCC AR6 as defined in Iturbide et al. (2020) and implemented using regionmask.
[11]:
all_inbuilt_weights = tas.get_scm_timeseries_weights(
regions=WEIGHTS_FUNCTIONS_WITHOUT_AREA_WEIGHTING.keys()
)
[12]:
# NBVAL_IGNORE_OUTPUT
plot_weights(all_inbuilt_weights)
<ipython-input-6-feff6056f082>:5: RuntimeWarning: More than 20 figures have been opened. Figures created through the pyplot interface (`matplotlib.pyplot.figure`) are retained until explicitly closed and may consume too much memory. (To control this warning, see the rcParam `figure.max_open_warning`).
ax = plt.figure().add_subplot(111)
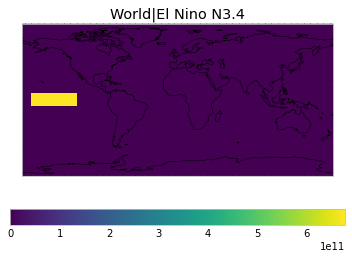
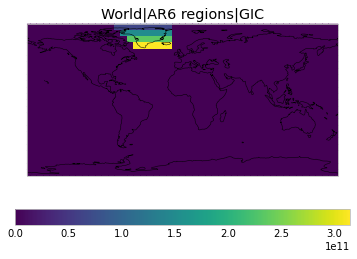
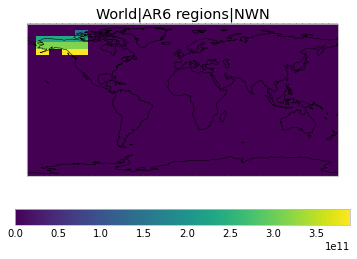
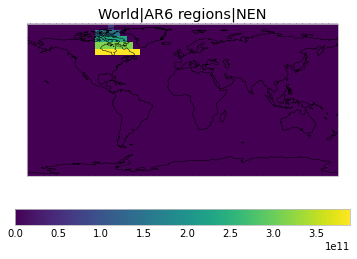
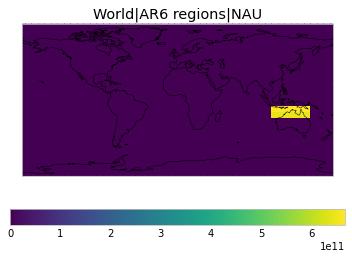
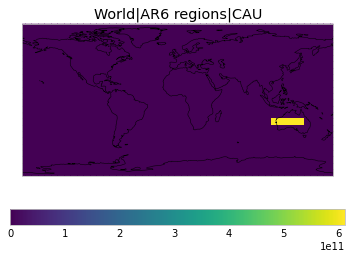
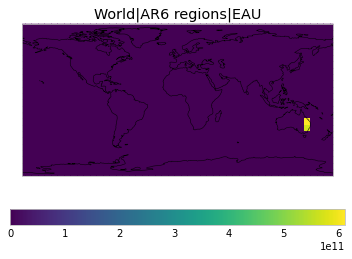
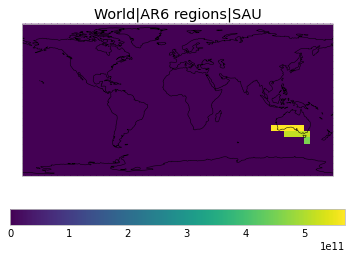
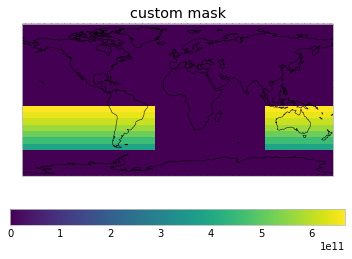
User-defined masks¶
As a user, you can also define masks. Simply add them to netcdf_scm.masks.MASKS and then use them in your get_scm_cubes call.
[13]:
WEIGHTS_FUNCTIONS_WITHOUT_AREA_WEIGHTING["custom mask"] = get_weights_for_area(
-60, 100, -10, 330
)
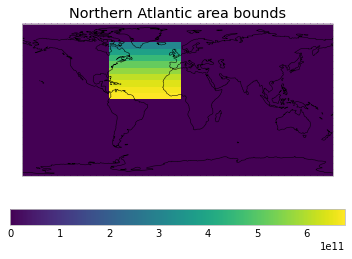
WEIGHTS_FUNCTIONS_WITHOUT_AREA_WEIGHTING[
"Northern Atlantic area bounds"
] = get_weights_for_area(0, -80, 65, 0)
[14]:
custom_weights = tas.get_scm_timeseries_weights(
regions=[
"World|El Nino N3.4",
"custom mask",
"World|Land",
"Northern Atlantic area bounds",
]
)
[15]:
plot_weights(custom_weights)